- Ironwood Ridge High School
- The Nighthawk Phylogeny Project
Manno, Theodore
Page Navigation
- About the Teacher
- Contact Info
- Bioscience I
- Bioscience II
- Bioscience Internship
- Biology (General)
- Nighthawk's Nest Biotechnology News
- Laboratory Gallery
- Classroom Gallery
- Nighthawk Chess Collective
- Nighthawk Origami & Tangram
- The Nighthawk Phylogeny Project
- Nighthawk Proteomics Institute
- Biostatistics @ The Nighthawk's Nest
- Ironwood Writer's Workshop
- The Ridgewood Methods
- Nighthawk Biotechnology Class Mascots
- Nighthawk Biotechnology In Action--Video Montage
-
THE NIGHTHAWK PHYLOGENY PROJECT
Awesome.
Phylogenetics is the study of the evolutionary history and relationships among or within groups of organisms. In the course of laboratory activities in Bioscience 2 and Bioscience-Internship, students are participating in THE NIGHTHAWK PHYLOGENY PROJECT, a long-term endeavor to collect, maintain, and memorialize DNA from leaf samples of every species of plant life in the garden areas on campus. Students extract and sequence DNA from their field-collected samples and determine their evolutionary relationships from these data with a sophisticated computer program. The result is a phylogenetic "tree" with new "branches" from yearly additions and data revisions.
Our renewed focus on bioinformatics--an interdisciplinary field which combines biology, the physical sciences, computer and information science, and statistics to analyze and interpret biological data--leads to students who are prepared for postsecondary study and the challenges of a dynamic contemporary landscape for future biotechnicians. Students can apply the skills they learn from this project to various in vitro and in silico analyses (e.g., revisions of current taxonomy, evaluating recent studies of mutation and evolution of COVID-19).
Students collect and preserve leaf samples from campus outdoor areas or common grocery store items (such as spinach), and then conduct high-quality DNA extraction using reagent-grade materials and our top-notch, spacious lab facilities.

After cataloging their samples in a DNA library, students use a thermal cycler to conduct a Polymerase Chain Reaction (PCR), a technique recently made well-known to non-scientists as the gold standard for diagnostic COVID-19 testing, and amplify (multiply) a particular gene from their DNA (using forwards and backwards rbcL primer).

Students clean their PCR product, elute the result in ultra-pure MilliQ water, and test their sample for purity with a Thermofisher Nanodrop. A label indicating the level of sample purity is generated for our refrigerated DNA library in the laboratory and for the microcentrifuge tube that is submitted for sequencing.

Students also confirm the presence of PCR amplicons (product) in their sample by conducting a gel electrophoresis procedure, where pieces of DNA are separated as they are "run" through an agarose gel in an electrode-containing apparatus. Having mixed, poured, and prepared all materials the day before, students vortex their samples, add gel loading dye to the "wells" in their agarose gel, fill their apparatus with TAE buffer, and conclude by staining the DNA in their gel with "Fast-Blast" dye. Gel Viewer Software generates images of where and how far the pieces of DNA that were created by the PCR reaction traveled, showing that the PCR amplified the gene of interest.

Once the sample is clean and pure enough for Sanger sequencing, students submit it to a specialized lab at the University of Arizona Genetics Core, BIO5 Institute. A few days later, students receive files with FASTA code for analysis.
EXAMPLE SEQUENCE
>X0z_lvs_00X0_rbcL_5420769_00X.ab1
TGGGGNGANAGCGGGTGTAAGAGTACAAATTGGACTT
ATTATACTCCTGGANTACGCAAACCAAAGGATACTGCA
TATCTTGGCAGCATTCCGAGTAACTCCTCAACCTGGAG
TTCCGCCTGAAGAAGCAGGGGCCGCGGTAGCTGCCG
AATCTTCTACTGGTACATGGACAACCGTGTGGACCGAT
GGACTTACCANTCTTGATCGTTACAANGGACGATGCTA
CCACATCGACCCGGNTCCTGGAGAAGCNGATCAATAT
ATCTGTTATGTAGCTTACCCTTTANACCTTTTCGAAGAA
GGNTCTTTACTAACATGTTTACTTCCATTGTAGGAAATG
TATTNGGATTCAAAGCCCTACGTGCTCTACNCTGGAAG
ATCTACNAATCCCTNCTGCTTATNTTAANACTTTCCANG
GNCCGCCTCACGGGACCAAGTTGAGAGAGATAAATTG
AACAAGTATGGTCGTCCCCTGTTGGGATGTACTNTTAA
ACTAAATTGGGATTATCTGCTAAANACTACGGTNGAGC
NNTTNATGANTGTCTTCGTGGGGGG
Students generate and examine spectrograms of their FASTA sequences and amino acid translations using 4-Peaks software, and then upload their FASTA code to a common file with every known DNA sequence from the project.
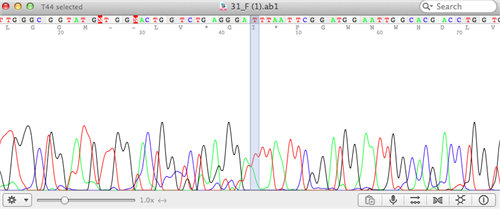


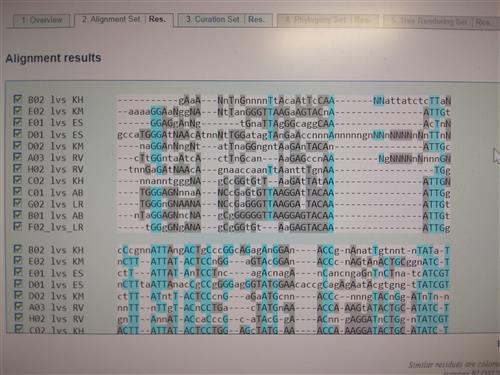
Using a computer program, students align all of the code from their amplified regions of interest with an algorithm. Students also identify the species from which their samples came by using a searchable database called NCBI-BLAST.



Students then generate a model phylogeny for the recorded species and analyze statistics to determine how likely their model is to apply to current evolutionary scenarios.

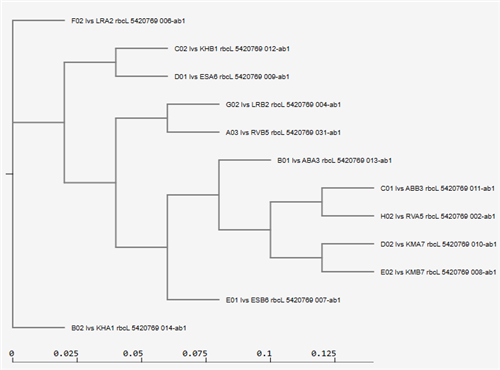
Students sometimes use other computer programs (e.g., Sequence Manipulation Site-SMS, JMOL, RCSB Protein Data Bank-PDB) to translate their DNA sequence into amino acid (protein) FASTA code and to generate models of 3D protein structures that are coded for by their sequenced DNA. THE NIGHTHAWK PHYLOGENY PROJECT hopes to have rbcL-amplified DNA sequences for all plant species on campus in a few years, with a phylogenetic tree that integrates their apparent genetic relatedness based on our generated models.
As a capstone to their participation in THE NIGHTHAWK PHYLOGENY PROJECT, students create a poster and/or full-length research paper that communicates the fundamental conclusions of their study.
Come join us! IRHS students have a unique opportunity to generate data that will be referred to for years to come. We are the future. And the future is now.

